P901 A genetic analysis of familial aggregation in Inflammatory Bowel Disease multiplex families
Jans, D.(1)*;Lee, H.S.(2);Ferrante, M.(3,4);Vermeire, S.(3,4);Cleynen, I.(1);
(1)KU Leuven, Department of Human Genetics- Laboratory for Complex Genetics, Leuven, Belgium;(2)University of Ulsan College of Medicine, Department of Biochemistry and Molecular Biology, Seoul, Korea- Republic Of;(3)KU Leuven, Department of Chronic Diseases- Metabolism and Ageing CHROMETA, Leuven, Belgium;(4)University Hospitals Leuven, Department of Gastroenterology and Hepatology, Leuven, Belgium;
Background
Relatives of patients with inflammatory bowel disease (IBD) have a higher risk to develop IBD than the general population. In some families, even a remarkably high prevalence of disease is found. Here, we aim to determine the role of the known IBD risk loci in familial aggregation of disease, as well as to study if there are common variants specific to familial IBD.
Methods
Imputed immunochip genotypes of 55 families [157 Crohn’s disease (CD), 29 ulcerative colitis (UC) and 138 unaffected relatives] that have at least three affected, first-degree relatives were used. A sporadic dataset (1705 CD, 917 UC, 873 controls) was used for comparison. For each individual, a polygenic risk score (PRS) was calculated with PRSice based on the IBD summary statistics of de Lange et al (2017), a p-value threshold ≤0.01 and MAF≥0.01. The mean PRS of all relatives of one family is denoted the familial PRS. Family-based association analyses were performed with SAIGE. The significance threshold for association used was p<1e-4.
Results
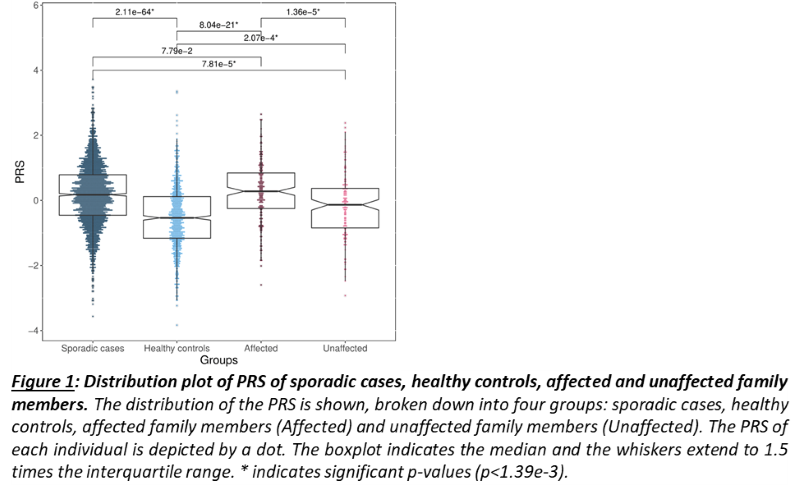
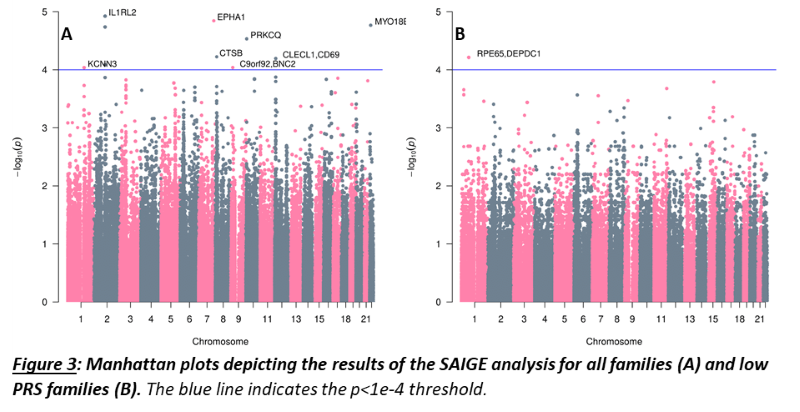
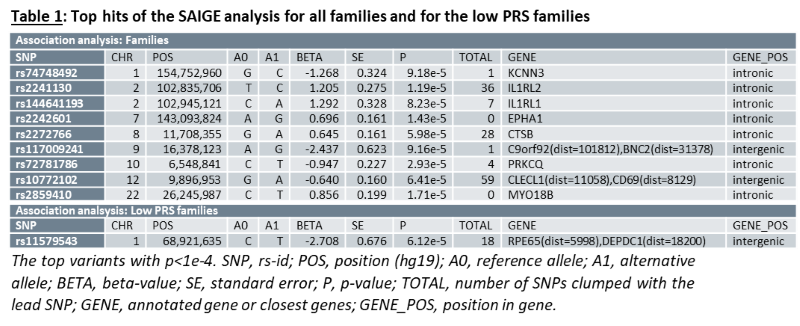
As a group, unaffected relatives have a lower PRS than affected relatives (p=1.36e-5, β(se)=-0.60(0.14)), but still higher than unrelated controls (p=2.07e-4, β(se)=0.39(0.11))(fig 1). Twenty-four families have a familial PRS that is higher than the mean PRS of sporadic cases, and seven families have a familial PRS that is lower than the mean PRS of unrelated controls, the low PRS families (fig 2A). In most families, the affected members have a higher mean PRS than the unaffected members (fig 2B). This relationship however is sometimes reversed, especially in the low PRS families. SAIGE found nine variants to be associated with familial IBD (fig 3A, table 1). Two independently associated variants, rs2241130 in IL1RL2 (p=1.19 e-5, β(se)=1.21(0.28)) and rs144641193 in IL1RL1 (p=8.23e-5, β(se)=1.29(0.33)) reside in a known IBD locus (IL18RAP). Some other identified variants were also located in genes previously implicated in IBD, either through GWAS [rs72781786 in PRKCQ (p=2.93e-5, β(se)= -0.95(0.23))], or via functional studies (rs2242601 in EPHA1 and rs2272766 in CTSB). A restricted analysis of the low PRS families identified only one significant variant: rs11579543, located between RPE65 and DEPDC1 (p=6.12e-5, β(se)=-2.71(0.68))(fig 3B, table 1).
Conclusion
Our analysis indicates that common risk variants play an important role in familial aggregation of disease in many multiplex families. Some families however have a very low polygenic risk, indicating shared environmental factors might be relatively more important, or genetic factors not captured by the score, e.g. rare variants, may have contributed. Also, a few loci are specifically implicated in familial IBD, some related to the immune system, e.g. CLECL1 and CD69.