OP03 Highly stable epigenome-wide peripheral blood DNA methylation signatures accurately predict endoscopic response to adalimumab, vedolizumab and ustekinumab in Crohn’s disease patients: The EPIC-CD study
JoustraMD., V.(1)*;Li Yim, A.(2,3);Hageman, I.(2);Levin, E.(4);Noble, A.(5);Chapman, T.(6);McGregor, C.(6);Adams, A.(6);Satsangi, J.(6);de Jonge, W.(2);Henneman, P.(3);D'Haens, G.(1);
(1)Amsterdam UMC location AMC, Gastroenterology and Hepatology, Amsterdam, The Netherlands;(2)Amsterdam UMC location AMC, Tytgat institute for Liver and Intestinal Research, Amsterdam, The Netherlands;(3)Amsterdam UMC location AMC, Genome Diagnostics Laboratory- Department of Clinical Genetics, Amsterdam, The Netherlands;(4)Horaizon, bv, Delft, The Netherlands;(5)Oxford University - Hospitals NHS Foundation Trust - John Radcliffe Hospital, Translational Gastroenterology Unit - NIHR Oxford Biomedical Research Centre, Oxford, United Kingdom;(6)Oxford University- Hospitals NHS Foundation Trust- John Radcliffe Hospital, Translational Gastroenterology Unit- NIHR Oxford Biomedical Research Centre, Oxford, United Kingdom; on behalf of the EPIC-CD Consortium
Background
Despite the proven efficacy of biological treatments in Crohn’s disease (CD), many patients fail to respond or lose response over time. Therefore, predictive biomarkers for treatment efficacy would be of extreme value. Previous epigenome-wide association studies associated differential DNA methylation with CD-specific phenotypes, suggesting a potential use in classification and prediction of response to treatment.
Methods
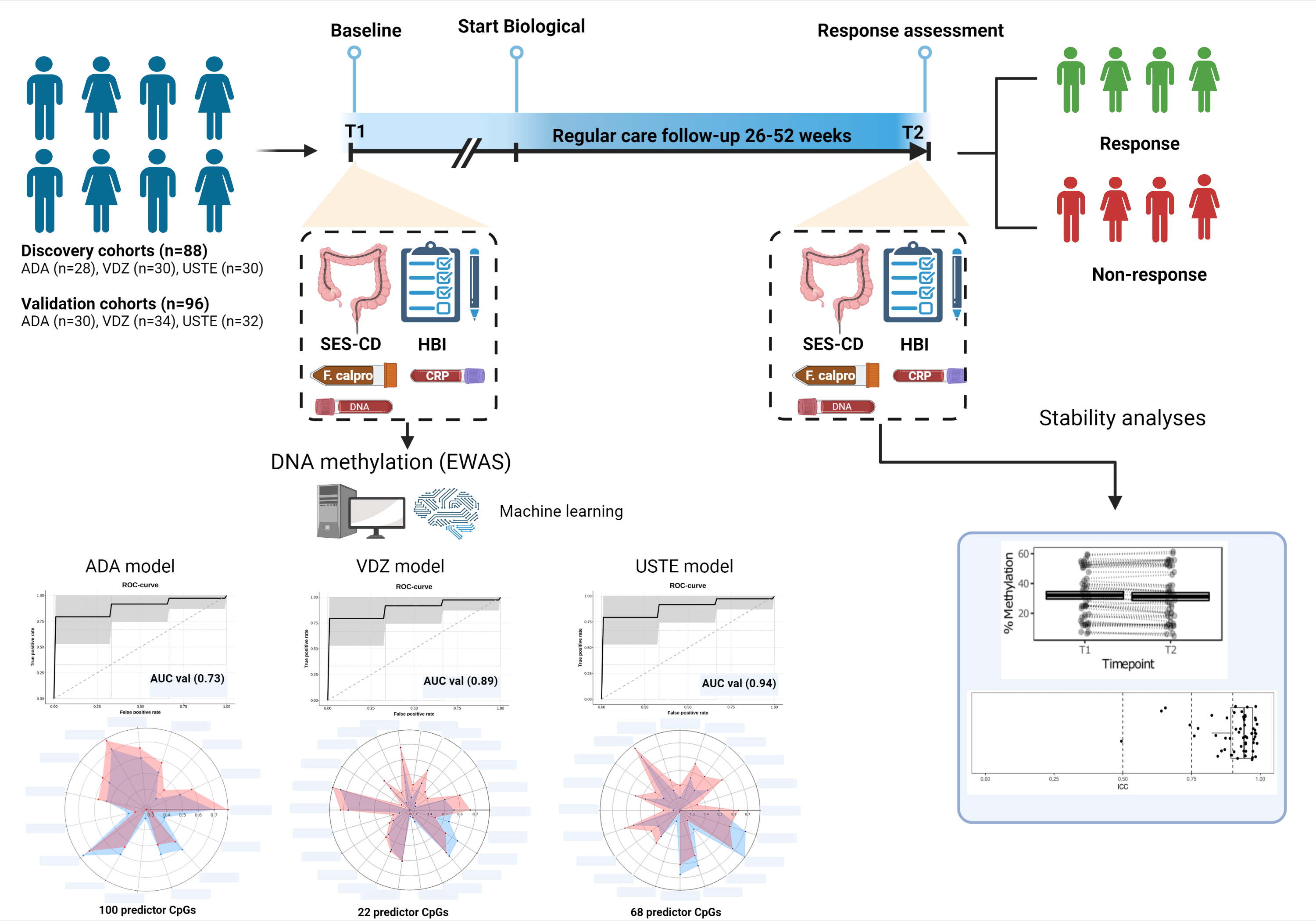
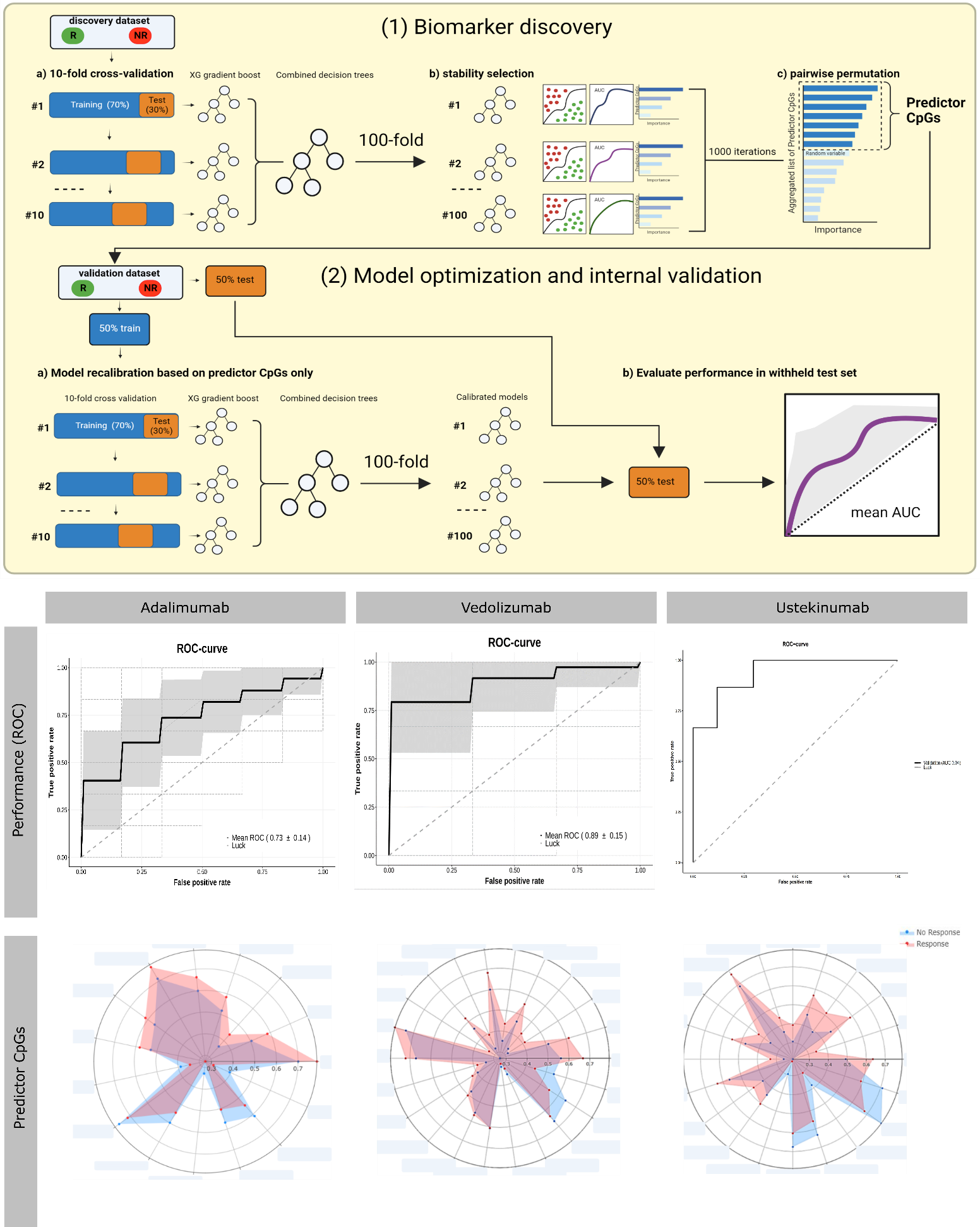
We prospectively collected and measured longitudinal peripheral blood DNA methylation profiles of 184 adult CD patients prior to (T1) and after a median of 28 weeks (T2) following biological treatment with Adalimumab (ADA), Vedolizumab (VEDO) or Ustekinumab (USTE) in a discovery (n=88) and independently collected internal validation cohort (n=96) using the Illumina EPIC BeadChip array. Response (R) was defined as the combination of endoscopic response (≥50% reduction in SES-CD score) and steroid-free clinical response (≥3 point drop in HBI or HBI ≤4 AND no systemic steroids) and/or biochemical response (≥50% reduction in C-reactive protein (CRP) and fecal calprotectin or a CRP ≤5 g/mL and fecal calprotectin ≤250 µg/g). Biomarker identification and classification analyses were performed using stability selection gradient boosting on samples taken at T1 whereas samples taken at T2 and intraclass correlation (ICC) data were used to assess long-term stability of our identified CpG loci1.
Results
A total of 58 ADA-patients (NR=29, NNR=29), 64 VEDO-patients (NR=36, NNR=28) and 62 USTE-patients (NR=30, NNR=32) were included. Prior to treatment, at T1, we identified distinct panels of 100 ADA-, 22 VEDO- and 68 USTE-associated CpG loci that, in combination, predict clinical- and endoscopic response with high accuracy (AUC ADA=0.73, VEDO=0.89 and USTE=0.94) upon validation. Notably, for these CpG loci, methylation levels did not significantly differ between T1 and T2, implicating stability during both induction and maintenance treatment, irrespective of inflammatory status and therapeutic intervention. In addition, the majority of these CpG loci (>60%) demonstrated long-term hyper stability (ICC-values ≥0.90). Furthermore, genes annotated to the CpGs of interest suggest drug specific involvement in TNF-signaling, endothelial cell-cell adhesion, integrin dependent T-cell homing, the innate immune system and Th17/Treg differentiation, corroborating to the mode of action of each drug.

Conclusion
Here, we report on 3 validated panels of highly stable, epigenetic biomarkers that predict clinical and endoscopic response in CD patients treated with ADA, VEDO or USTE. Additional external- and clinical validation as part of EPIC-CD and the OMICROHN clinical trial are currently ongoing.