P027 Epithelial cells of patients with ulcerative colitis do not show an increased sensitivity after microbiota stimulation compared to non-IBD controls
Arnauts, K.(1,2);Lapierre, C.(1);Verstockt, B.(1,3);Verstockt, S.(1);Sudhakar, P.(1);Vermeire, S.(1,3);Verfaillie, C.(2);Sabino, J.(1,3);Ferrante, M.(1,3);
(1)KU Leuven, TARGID- Department of Chronic Diseases- Metabolism & Ageing, Leuven, Belgium;(2)KU Leuven, Stem Cell Institute Leuven- Department of Development and Regeneration, Leuven, Belgium;(3)KU Leuven, Department of Gastroenterology and Hepatology- University Hospitals Leuven, Leuven, Belgium
Background
Alterations in the intestinal microbiota play a pivotal role in the pathogenesis of Inflammatory Bowel Diseases (IBD). Although there is a lot of interest in restoring dysbiosis, the effects of microbial alterations are not fully understood. In addition, it is known that epithelial cells from IBD patients maintain intrinsic defects1. For that reason, our aim was to unravel if epithelial cells of UC patients are more sensitive towards microbiota stimulation, compared to non-IBD controls.
Methods
Intestinal organoids of UC patients (n=8) and non-IBD controls (n=8) were grown as monolayers on Transwell inserts. Upon confluency (evaluated by transepithelial electrical resistance (TEER)), monolayers were stimulated for 24 hours with TNF-α (100 ng/ml), IL-1β (20 ng/ml) and Flagellin (1 µg/ml) to mimic inflammation. Fresh fecal samples of a selected donor (n=1, high microbial cell count and presence of selected phyla2) and UC patients (n=3, endoscopic sub-mayo ≥2) were filtered and stored in 0.9% NaCl. Monolayers were stimulated for 6 hours with 3.108 microbial cells (cell count by Flow Cytometry). RNA sequencing was performed by Truseq for Illumina. Differentially expressed genes (DEG) were studied by DESeq2 (FDR <0.05).
Results
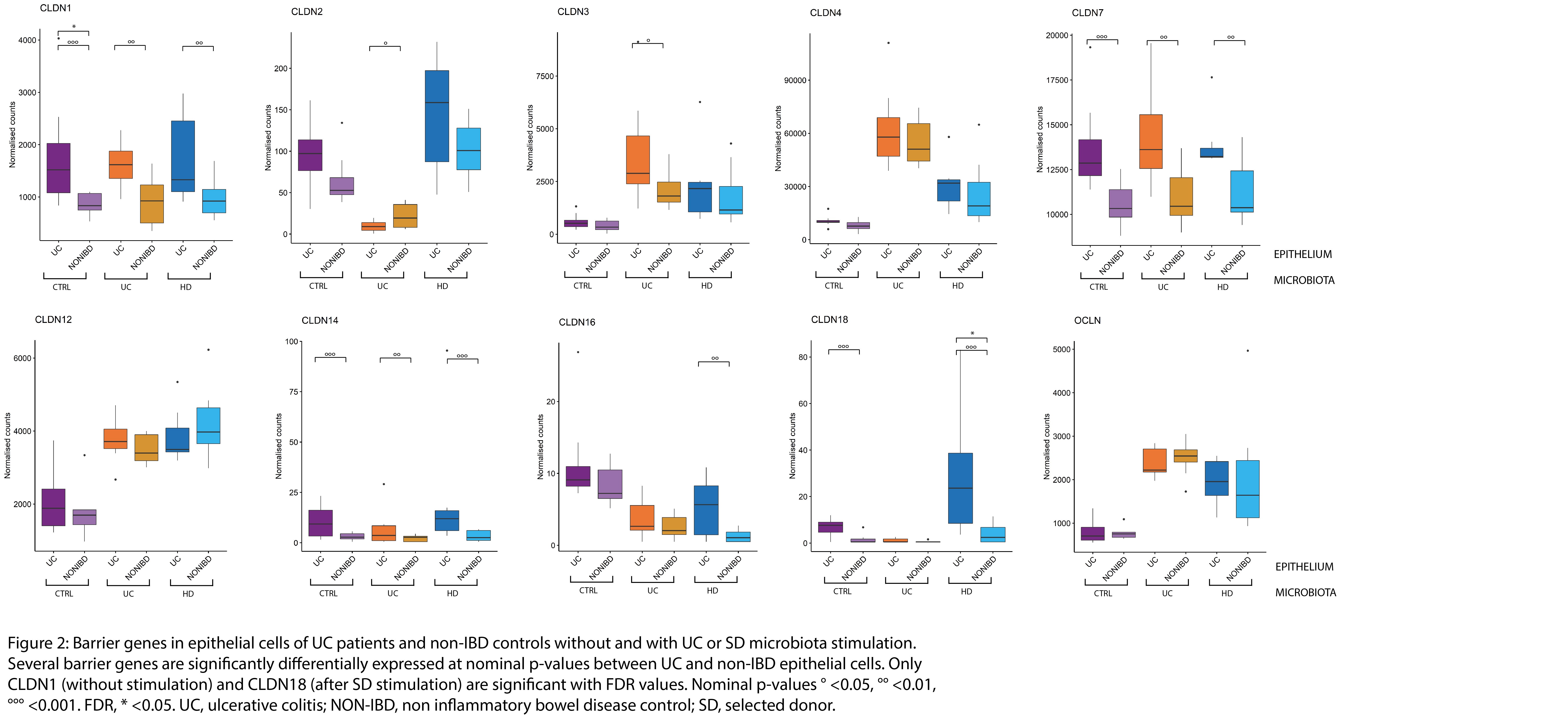
Although TEER measurements indicated a higher epithelial cell permeability upon UC microbiota stimulation in UC patients compared to non-IBD controls (p=0.038; Mann-Whitney; Figure 1), we could not confirm this distinct response based on RNA sequencing data at principal component analysis (PCA). Several epithelial barrier genes were significantly upregulated between UC and non-IBD epithelium at nominal p-value, while only CLDN1 and 18 were significant for FDR <0.05 (Figure 2). Clustering on PCA was driven by microbial treatment and not by epithelial origin (Figure 3). Inflamed monolayers of UC patients showed different baseline characteristics (129 DEG; e.g. HLA-G, MUC2, CLDN1, IL23A, PARP8; Figure 4A), but did not propagate in a different response upon microbiota exposure compared to non-IBD controls. Treatment with microbiota of UC patients (23 DEG; e.g. PARP9, TGFBI, ANXA13) or the selected donor (58 DEG; e.g. CCL5, CLDN18, TGFBI) only induced minor differences between epithelial cell types (Figure 4B).
Conclusion
We observed no different response in epithelial cells of UC patients towards microbiota stimulation compared to non-IBD epithelial cells on transcriptomic level. Further validation on barrier integrity is needed. We observed no indications that microbial treatment would be less beneficial to UC patients, based on the epithelial cell response. Addition of (patient specific) immune cells will contribute to unraveling host-microbiota interactions in IBD patients.
1.Dotti,Gut,2017
2.Vermeire,JCC,2015