P106 Identification of urine and serum diagnostic biomarkers of inflammatory bowel disease using a proteomic approach
Baldan-Martin, M.(1)*;Azkargorta, M.(2);Lloro, I.(2);Soleto Fernández, I.(1);Orejudo, M.(1);Ramírez, C.(1);García, S.(1);Mercado, J.(1);Riestra, S.(3);Rivero, M.(4);Gutiérrez, A.(5);Rodríguez-Lago, I.(6);Fernández-Salazar, L.(7);Ceballos, D.(8);Benítez, J.M.(9);Aguas, M.(10);Bastón-Rey, I.(11);Bermejo, F.(12);Casanova, M.J.(1);Lorente, R.(13);Ber, Y.(14);Royo, V.(15);Esteve, M.(16);Elortza, F.(2);Gisbert, J.P.(1);Chaparro, M.(1);
(1)Hospital Universitario de La Princesa- Instituto de Investigación Sanitaria Princesa IIS-Princesa- Universidad Autónoma de Madrid UAM- and Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas CIBERehd, Gastroenterology Unit, Madrid, Spain;(2)Proteomics Platform- CIC bioGUNE- BRTA Basque Research & Technology Alliance- CIBERehd- ProteoRed-ISCIII, Proteomics Platform, Derio, Spain;(3)Hospital Universitario Central de Asturias and Instituto de Investigación Sanitaria del Principado de Asturias ISPA, Gastroenterology Unit, Oviedo, Spain;(4)Hospital Universitario Marqués de Valdecilla and IDIVAL, Gastroenterology Unit, Santander, Spain;(5)Hospital General Universitario de Alicante- ISABIAL and CIBERehd, Gastroenterology Unit, Alicante, Spain;(6)Hospital Galdakao-Usansolo, Gastroenterology Unit, Vizcaya, Spain;(7)Hospital Clínico Universitario de Valladolid, Gastroenterology Unit, Valladolid, Spain;(8)Hospital Universitario de Gran Canaria Dr. Negrín, Gastroenterology Unit, Las Palmas de Gran Canaria, Spain;(9)Hospital Universitario Reina Sofía and IMIBIC, Gastroenterology Unit, Córdoba, Spain;(10)Health Research Institute IISLaFe and Hospital Universitari i Politecnic La Fe, Gastroenterology Unit, Valencia, Spain;(11)Hospital Clínico Universitario de Santiago de Compostela, Gastroenterology Unit, Santiago de Compostela, Spain;(12)Hospital Universitario de Fuenlabrada and IDIPAZ, Gastroenterology Unit, Madrid, Spain;(13)Hospital General Universitario de Ciudad Real, Gastroenterology Unit, Ciudad Real, Spain;(14)Hospital San Jorge, Gastroenterology Unit, Huesca, Spain;(15)Hospital Universitari Son Espases, Gastroenterology Unit, Palma de Mallorca, Spain;(16)Hospital Universitari Mutua Terrasa and Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas CIBERehd, Gastroenterology Unit, Terrasa, Spain;
Background
Inflammatory bowel diseases (IBD) are chronic, heterogeneous, and inflammatory conditions mainly affecting the gastrointestinal tract. Currently, endoscopy is the gold standard test for assessing mucosal activity and healing in clinical practice; however, it is a costly, time-consuming, invasive, and uncomfortable procedure for patients. There is, therefore, a need for sensitive, specific, fast and non-invasive biomarkers for the diagnosis of IBD.
Methods
Label-free quantification by nanoscale liquid chromatography coupled to tandem mass spectrometry (nLC MS/MS) was performed to profile the urinary and serum proteomes of 100 patients with newly diagnosed IBD, before starting any treatment [50 patients with Crohn´s disease (CD) and 50 patients with ulcerative colitis (UC)] and 50 healthy controls (HC). Data Mining and Pattern Recognition techniques were used to identify hidden relationships that are not detectable by using the classical and most commonly used lineal classifiers; and thus, identifying potential markers able to discriminate the studied cohorts. Finally, we applied Ingenuity Pathway Analysis (IPA) to analyze the pathways and functions of differentially expressed proteins.
Results
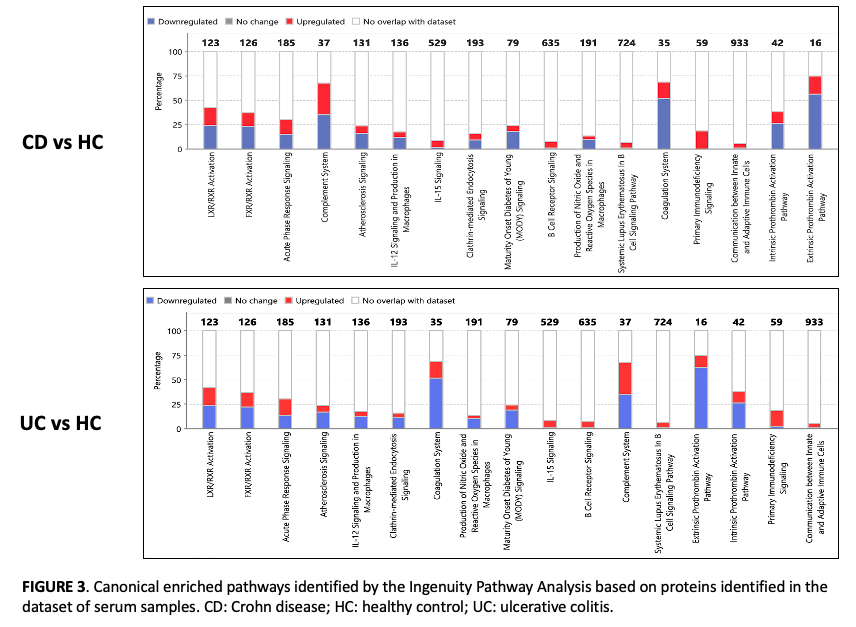
Serum proteomics results revealed 45 differentially expressed proteins in the comparison between UC and HC groups, 32 proteins significantly expressed in CD versus HC, and 12 proteins in CD compared with UC. In urine samples, 110 proteins were significantly changed in UC versus HC, 50 proteins were differentially expressed between CD and HC, and a total of 31 proteins were significantly changed in CD compared with UC patients. Receiver operating characteristic curve (ROC) analysis found multiple proteins with high area under the curve values, up to 0.94, indicating that these serum and urine proteins are of value as new non-invasive diagnostic classifiers of IBD patients. For each study comparative, the 5 most significant classifiers were selected (Figures 1 and 2). IPA revealed multiple signaling pathways, including prothrombin activation, acute phase response signaling, complement and coagulation system and liver X receptor/retinoid X receptor (LXR/RXR) activation, altered in IBD patients compared to HC (p-value < 0.05) (Figures 3 and 4). 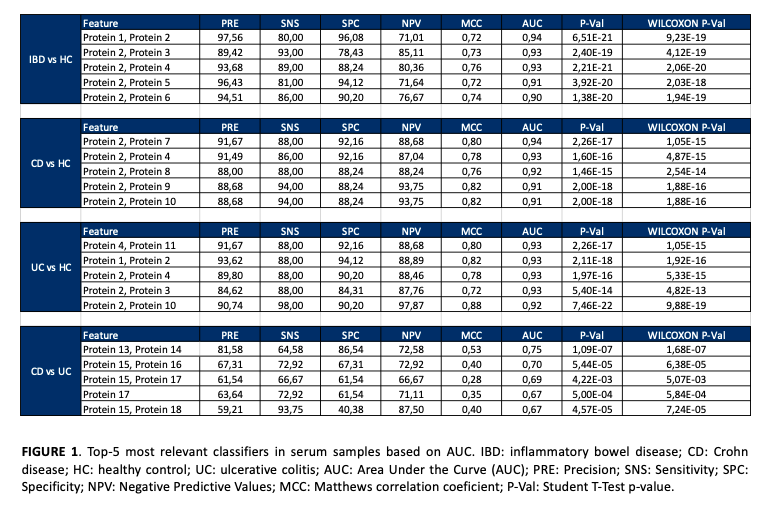


Conclusion
Our findings indicate that analysis of the urine and serum proteome using nLC MS/MS is a feasible approach for biomarker discovery. We identified several serum and urine proteins that could serve as new non-invasive markers for the diagnosis of IBD patients after further validation. After bioinformatic analyses, we found multiple proteins that may play important roles in the pathogenesis of IBD.


