P154 A novel serum protein signature as biomarker for Inflammatory Bowel Disease: A diagnostic performance and prediction modelling study using data from two independent inception cohorts
Bazov, I.(1);Kruse, R.(2);Bergemalm, D.(3);Eriksson, C.(3);Hedin, C.R.(4,5);Carlson, M.(6);van Nieuwenhoven, M.(3);Keita, Å.V.(7);Magnusson, M.K.(8);Almer, S.(4,9);Strid, H.(10);Bache-Wiig Mathisen, C.(11,12);Bengtsson, M.B.(13);Bergene Aabrekk, T.(12,13);Medhus, A.W.(11,12);Detlie, T.E.(14,15);Frigstad, S.O.(16);Huppertz-Hauss, G.(17);Opheim, R.(11,18);Ricanek, P.(14,19);Kristensen, V.A.(11,20);Salihovic, S.(21);D'Amato, M.(22,23); Öhman, L.(8);Söderholm, J.D.(7);Lindqvist, C.M.(24);Repsilber, D.(1);HøivikMD- PhD, M.L.(11,12);Halfvarson, J.(3)*;
(1)Örebro University, School of Medical Sciences, Örebro, Sweden;(2)Örebro University, Department of Clinical Research Laboratory- Faculty of Medicine and Health, Örebro, Sweden;(3)Örebro University, Department of Gastroenterology- Faculty of Medicine and Health, Örebro, Sweden;(4)Karolinska Institutet, Department of Medicine- Solna, Stockholm, Sweden;(5)Karolinska University Hospital, Gastroenterology unit- Department of Gastroenterology- Dermatovenereology and Rheumatology-, Stockholm, Sweden;(6)Uppsala University, Department of Medical Sciences, Uppsala, Sweden;(7)Linköping University, Department of Biomedical and Clinical Sciences, Linköping, Sweden;(8)University of Gothenburg- Sahlgrenska Academy, Department of Microbiology and Immunology- Institute of Biomedicine, Gothenburg, Sweden;(9)Karolinska university hospital, Gastroenterology unit- Department of Gastroenterology- Dermatovenereology and Rheumatology, Stockholm, Sweden;(10)Södra Älvsborgs Hospital, Department of Internal Medicine, Borås, Sweden;(11)Oslo University Hospital, Department of Gastroenterology, Oslo, Norway;(12)University of Oslo, Institute of Clinical Medicine, Oslo, Norway;(13)Vestfold Hospital Trust, Department of Gastroenterology, Tønsberg, Norway;(14)Akershus University Hospital, Department of Gastroenterology, Lørenskog, Norway;(15)University of Oslo, Insititute of Clinical Medicine, Oslo, Norway;(16)Vestre Viken Hospital Trust- Bærum Hospital, Department of Internal Medicine, Bærum, Norway;(17)Telemark Hospital Trust- Skien, Department of Gastroenterology, Skien, Norway;(18)University of Oslo, Institute of Health and Society, Oslo, Norway;(19)Lovisenberg Diaconal Hospital, Department of Gastroenterology, Oslo, Norway;(20)Lovisenberg Diaconal Hospital, Unger-Vetlesen Institute, Oslo, Norway;(21)Örebro University, School of Medical Sciences- Inflammatory Response and Infection Susceptibility Centre, Örebro, Norway;(22)Karolinska Institutet, Clinical Epidemiology Division- Department of Medicine Solna, Stockholm, Sweden;(23)IKERBASQUE- Basque Foundation for Science- Bilbao, Gastrointestinal Genetics Lab- CIC bioGUNE - BRTA, Bilbao, Spain;(24)Örebro University, School of Medical Sciences- Faculty of Medicine and Health, Örebro, Sweden; SIC-IBD and IBSEN III
Background
The diagnosis of inflammatory bowel disease (IBD) is often delayed since existing biomarkers in blood are imprecise and faecal tests are poorly accepted by patients. Therefore, we aimed to identify a diagnostic serum protein signature and validate its performance and utility.
Methods
Proximity extension immunoassay (PEA) methodology (Olink Proteomics, Uppsala, Sweden) was used to measure 184 immune and inflammatory markers in serum samples from adult patients with suspected IBD in the Swedish inception cohort SIC IBD (discovery cohort, n=451) and the Norwegian population-based inception cohort IBSEN III (validation cohort, n=554). Samples were obtained at presentation, i.e., almost all patients were treatment naïve. Diagnostic assessment was performed following the ECCO guidelines. Supervised machine learning with smoothly clipped absolute deviation (SCAD) regularised logistic regression and random forest (RF) models were employed in nested cross-validation to identify a diagnostic signature of IBD.
Results
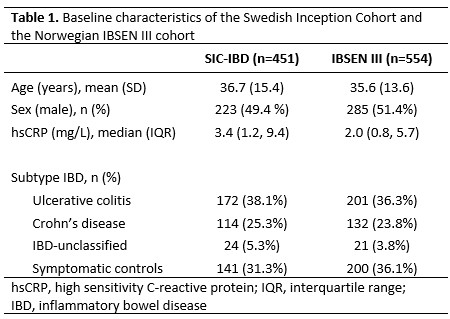
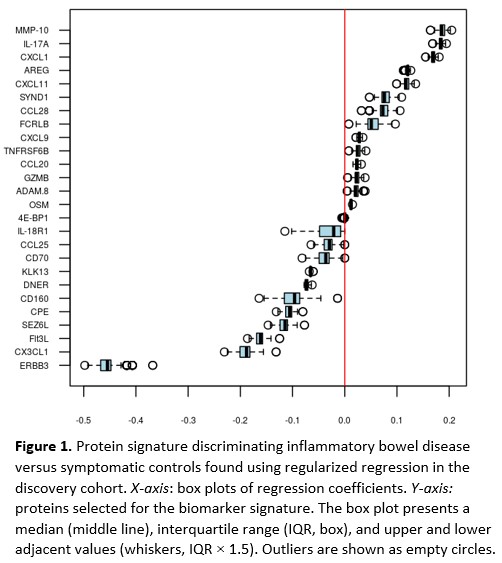
The discovery cohort included 310 IBD patients and 141 symptomatic controls without any discernible evidence of IBD, and the validation cohort included 354 IBD patients and 200 symptomatic controls (Table 1). The regularised regression identified a signature of 26 proteins that distinguished IBD from symptomatic controls with an area under the curve (AUC) of 0.78 in the discovery cohort. Contributions of individual proteins for the diagnostic signature are shown in Figure 1. Next, we validated the diagnostic capacity of the protein signature in the validation cohort (AUC=0.82) and compared it to high-sensitivity C-reactive protein (hsCRP) (Figure 2, AUC=0.69, p=1.1×10-7). The RF model yielded similar results. Only 422/554 patients in the validation cohort provided a stool sample. Among these patients, the diagnostic accuracy of the protein signature (AUC=0.82) and faecal Calprotectin (AUC=0.86) did not differ (p=0.07). A complex protein signature may preclude its introduction into clinical practice. Simplified models were therefore constructed using discovery cohort data and stepwise forward logistic regression with hsCRP as the null model. When applied to the validation cohort, the diagnostic accuracy of the shortest signature, comprising hsCRP and interleukin (IL)-17A (AUC=0.80), was superior to hsCRP (Figure 3, AUC=0.69, p=1.7×10-8).


Conclusion
A serum protein signature of hsCRP and IL-17A was superior to hsCRP in differentiating treatment-naïve IBD from symptomatic controls. Taking into account that many patients do not accept to provide faecal samples, we believe this signature has the potential to become an accessible and scalable test that can shorten the diagnostic delay in IBD.


