P386 Predictive models for assessing the response to ustekinumab in Crohn’s disease patients
Hubert, A.(1);Bottieau, J.(1);Toubeau, J.F.(1);Franchimont, D.(2,3);Vallée, F.(1);Liefferinckx, C.(2,3);
(1)University of Mons, Department of Electrical Engineering, Mons, Belgium;(2)Hôpital Erasme, Department of Gastroenterology, Brussels, Belgium;(3)Université libre de Bruxelles, Laboratory of experimental gastroenterology, Brussels, Belgium;
Background
The loss of response to biologics remains a clinical challenge for which the discovery of predictive factors is helpful. In this direction, exploiting tree-based models on top of logistic regression (LR) allows to uncover unexpected valuable predictors. The aim of the study is to develop performant predictions of ustekinumab (USK) response at one year of follow-up in a cohort of Crohn’s disease (CD) patients using benchmark logistic regression (LR) and tailored tree-based techniques, Random forest (RF) and Gradient Boosting Decision Trees (GBDT).
Methods
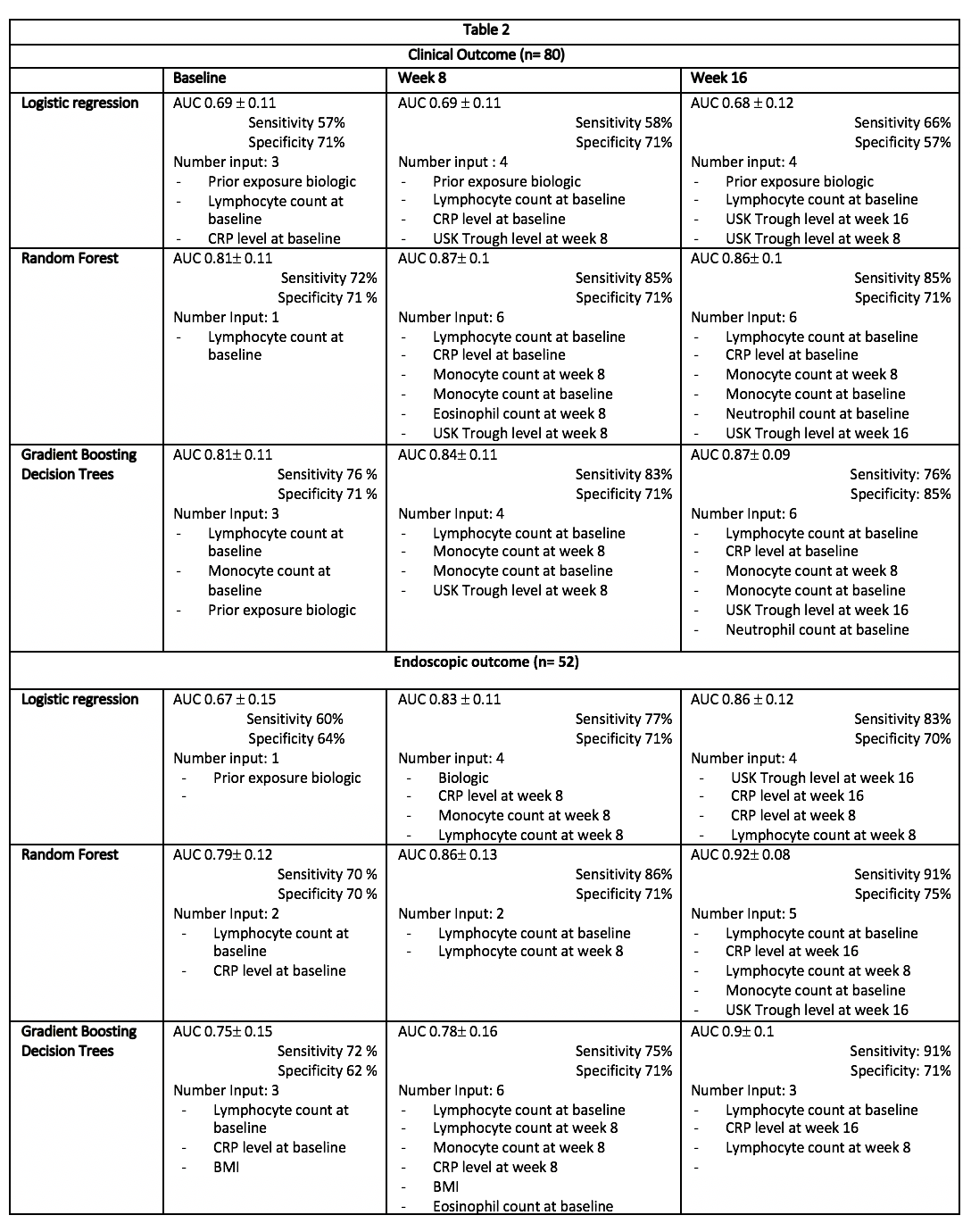
This is a retrospective study based on a cohort of 80 CD patients treated with USK. Inputs include demographic data, disease characteristics as well as pharmacokinetics, clinical and biological parameters at baseline, week 8 and 16. After one-year follow-up, both clinical outcome (achieved in case of USK maintenance without optimization, n=80) and endoscopic outcome (achieved in case of SES-CD<10, n=52) are evaluated. The inputs are firstly ranked via the p-values for LR method and the impurity criterion for the tree-based ensemble methods. Then, the optimal subset of inputs for both methods is selected following a sequential feature selection. Due to the limited size cohort, a nested cross-validation framework was applied to avoid bias and overfitting. The performance of the final models is evaluated using the area under the ROC curve (AUC) with standard deviation (SD), sensitivity (Se) and specificity (Sp). All the analyses are performed with Python 3.6 6 using the packages statsmodels and Scikit-learn..
Results
Input data characterizing the CD cohort are presented in Table 1 . At one-year follow-up, clinical and endoscopic outcomes were achieved in 56.2% (n=45/80) and 69% (n= 36/52), respectively. Table 2 summarizes the predictive model characteristics at the different timepoints. Whatever the outcome, predictive models exhibited an increased accuracy over the induction timepoints. At week 16, a multivariate LR allowed to predict endoscopic outcome with an AUC 0.86±0.12, Se 83% and Sp 70%. The explanatory variables were CRP level at week 8 and 16, lymphocyte count at week 8 and USK trough levels at week 16. The best model to predict endoscopic outcome was obtained by RF with an AUC of 0.92 ±0.08, Se 91% and Sp 75%. The key inputs were lymphocyte count at baseline.
Conclusion
This study highlighted the added value of applying tree-based models for the development of accurate predictive models. More specifically, the evolution of biological markers during induction as well as USK trough levels at week 16 are relevant factors to predict endoscopic outcome among CD patients treated with USK


