P718 Characterization of microbiota and metabolomic profile in human colonic surgical resections of UC patients
Bauset, C.(1);Carda-Diéguez, M.(2);Buetas, E.(2);Lis-López, L.(1);Navarro, F.(3);Ortiz-Masiá, D.(4);Calatayud, S.(1);Barrachina, M.D.(1);Mira, Á.(2);Cosín-Roger, J.(5);
(1)Universitat de Valencia, Pharmacology, Valencia, Spain;(2)FISABIO, Genomics and Health Department, Valencia, Spain;(3)Hospital de Manises, Colorectal surgery, Valencia, Spain;(4)Universitat de Valencia, Medicine, Valencia, Spain;(5)FISABIO, Unidad Mixta Facultad de Medicina-Hospital Dr. Peset, Valencia, Spain;
Background
Ulcerative colitis (UC) is characterized by a diffuse, continuous, and chronic inflammation of mucosa and submucosa layers in the colon whose etiology is still unknown. Intestinal microbiota dysbiosis and alterations in the metabolomic profile have been reported in mucosal biopsies from Inflammatory Bowel Disease (IBD) patients. We aim to characterize the microbiota composition and metabolomic profile in colonic resections of UC patients.
Methods
Colonic resections from UC (n=18) and non-IBD (n=20) patients were obtained. Microbiota identification, composition and classification was performed by 16S rRNA gene Illumina Miseq sequencing. The bioinformatic analysis of sequencing data was performed using constrained correspondence analysis (CCA) and non-parametric Wilcoxon test to compare species and genera proportions. Bacterial load was estimated by qPCR. Metabolomic analysis was performed using Nuclear Magnetic Resonance (NMR) to study polar metabolites and Gas-Chromatography or Liquid-Chromatography Mass-Spectrometry to study non-polar metabolites.
Results
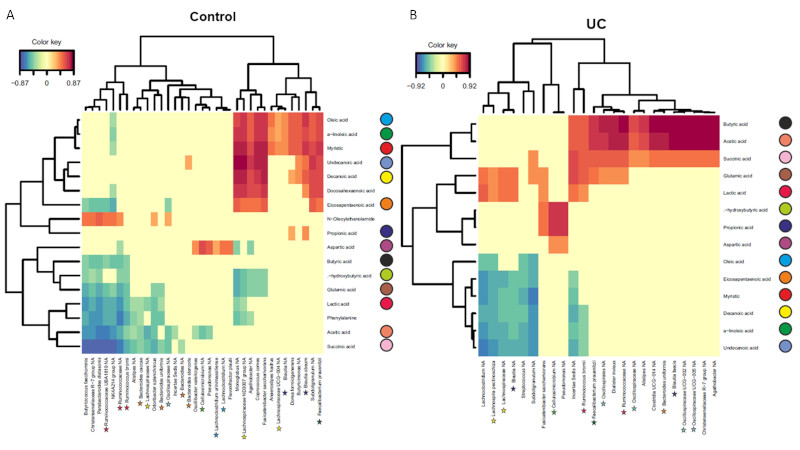
Microbiota analysis revealed differences at genus and species level between UC and non-IBD patients. Cellulosimicrobium was found at 10-fold higher levels in UC samples while Escherichia genus was 3.88 times more abundant in controls. A reduction in bacterial load was seen in UC samples. Levels of short-chain fatty acids (FA), medium-chain FA, long-chain FA, carboxylic acids and amino acids were obtained from the metabolomics analysis and a positive correlation between propionic, aspartic and hydroxybutyric acids and Cellulosimicrobium and Pseudomonas was found (Fig1B). Gut commensals such as unclassified Agathobacter, Blautia faecis, Oscillospiraceae or Faecalibacterium prausnitzii positively correlated with the abundance of butyric acid, acetic acid and succinic acid; meanwhile Subdoligranulum, Lachnospireace or unclassified Blautia negatively correlated with some medium or long chain fatty acids (Fig1B).
Fig1. Correlations between bacterial species and metabolites in (A) controls and (B) UC patients. Performed with mixOmics library in R.
Conclusion
Significant differences in microbiota composition and abundance are found in UC patients, which undergo a clear gut dysbiosis in colonic tissue. Correlation analysis suggests Cellulosimicrobium and Pseudomonas as sources of propionic, aspartic and hydroxybutyric acid. These results point at Cellulosimicrobium as a potential candidate in UC pathology.
- Posted in: Poster presentations: Microbiology 2022