J. Axelrad1, A. Hine2, J. Devlin2, P. Loke2, K. Cadwell2
1New York University School of Medicine, Gastroenterology, New York, USA, 2New York University School of Medicine, Microbiology, New York, USA
Background
Stool PCR pathogen panel assays have implicated enteric infection in nearly 30% of inflammatory bowel disease (IBD) flares, most commonly with Clostridioides difficile, Escherichia coli, and norovirus. Many questions remain about the interpretation of a positive stool PCR result, and the similar presentations of infection and flare present a clinical challenge. We aimed to characterise the gut microbiome during an infection with C. difficile, E. coli, or norovirus in patients with and without IBD to microbially differentiate patients with an enteric infection, flare of IBD, and flare of IBD complicated by a pathogen.
Methods
We performed a cross-sectional study of patients with a stool PCR panel positive for C. difficile, E. coli subtypes, norovirus, or negative for all pathogens during an episode of gastrointestinal symptoms from September 2018 to February 2019. Clinical data and remnant stool specimens were collected. Our primary outcome was microbiome signature using 16s bacterial rRNA gene sequencing derived from linear discriminant analysis effect size (LEfSe) to identify microbes associated with each pathogen and clinical scenario stratified by IBD.
Results
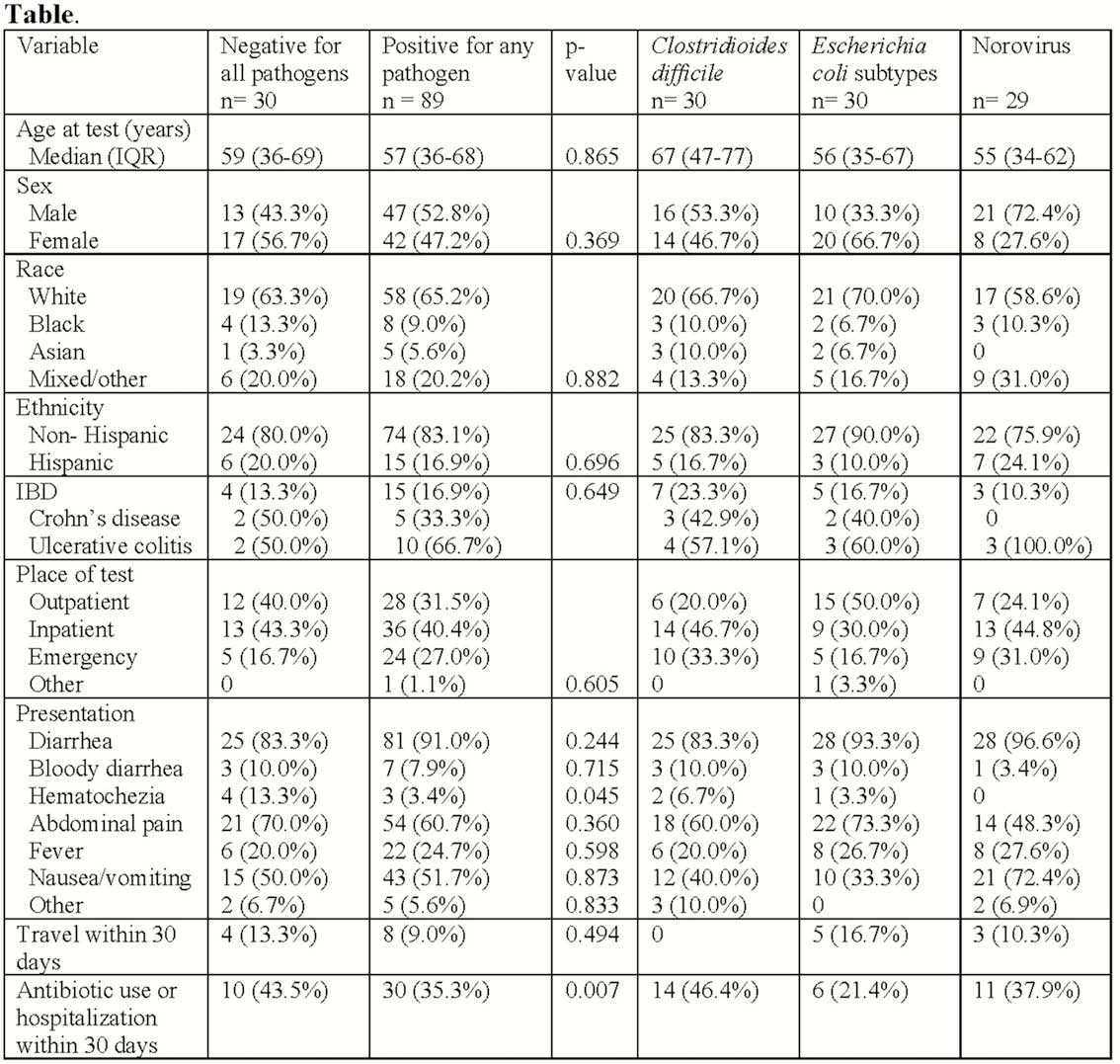
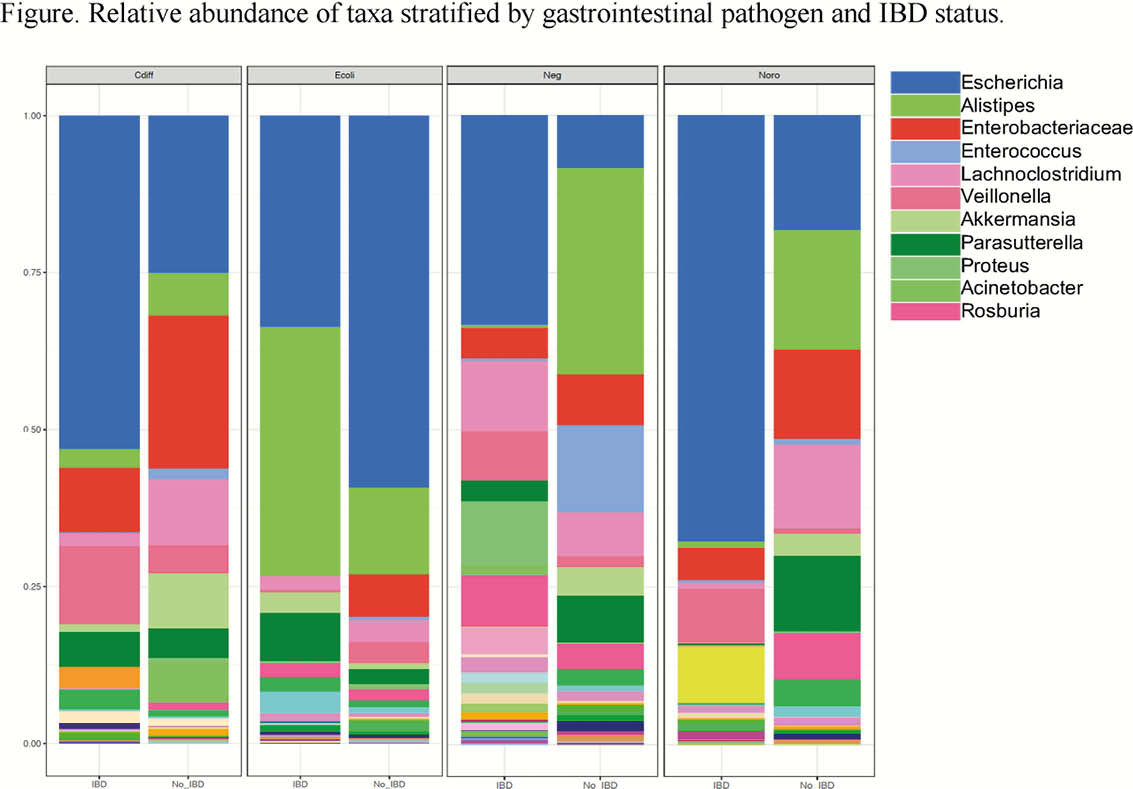
We identified 119 patients, 19 (16%) with IBD, with C. difficile (n = 30), an E. coli subtype (n = 30), norovirus (n = 29), or negative for all pathogens (n = 30; Table). Hematochezia on presentation and recent antibiotic or hospitalisation exposure was more common in pathogen negative patients compared with pathogen positive patients (p = 0.045, p = 0.007, respectively). Patients with IBD without a pathogen or with C. difficile tended to have higher serum biomarkers of inflammation (median CRP 24 and 28, respectively) compared with patients with an E. coli subtype (CRP 5) or norovirus (CRP 11). On LEfSe, patients with C. difficile were enriched in Enterobacteriaceae and Clostridioides, E. coli subtypes in Faecalibacterium, norovirus in Roseburia, and negative for a pathogen in Bacteroides. Stratifying by IBD and pathogen, there were several differentially abundant microbes denoting a specific gut microbial signature associated with each pathogen and IBD (Figure).
Conclusion
In this study of patients with C. difficile, E. coli, norovirus, or negative for pathogens during an episode of gastrointestinal symptoms, there were major differences in presentation and gut microbiota between patients with and without an enteric pathogen and IBD. These data demonstrate that gut microbiomes distinguish and differentially respond to pathogens. In patients with IBD, coupled with a unique microbiota, serum biomarkers of inflammation were lower in patients with E. coli or norovirus, suggesting a milder subset of IBD flare.




