P924 A unique gut microbiome signature can help differentiate Crohn`s Disease and isolated Terminal Ileitis
Raghunathan, N.(1)*;Joshi, V.(1);Patel, R.(1);Mekala, D.(1);Pal, P.(1);Banerjee, R.(1);
(1)Asian Institute of Gastroenterology, IBD Centre, Hyderabad, India;
Background
With increasing number of ileal intubations, isolated terminal ileal ulcers(TI) are often detected during colonoscopies. The etiology of these lesions have not been well-defined and differentiating TI from early Crohn’s Disease (CD) is a commonly encountered diagnostic challenge. CD is associated with significant dysbiosis; however, the gut microbiome of TI is poorly studied. We aimed to compare the gut microbial profile between CD and isolated non-specific TI.
Methods
Fecal samples were collected from treatment naïve patients (26 CD;13 TI) and healthy volunteers (29) without the use of antibiotics, probiotics, and prebiotics for at least 6 weeks. CD was confirmed by endoscopic and histological examination. TI was diagnosed based on isolated terminal ileal erosions/ulcers, normal small bowel endoscopy, non-specific inflammation with no granulomas/ cryptitis/crypt abscess on biopsy, and no history of NSAIDs. Fecal DNA was isolated and sequenced(16S-V4). Taxonomical classification was performed by Amplicon Sequence Variant approach and analysed by STAMP (Statistical Analysis of Metagenomic Profiles) using Welch`s T-Test. PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) was used for predictive metagenomics. Significant metabolic pathways were corrected by Storey FDR (q<0.05). Fecal Calprotectin (FCal) was measured using QuantOnCal rapid test.
Results
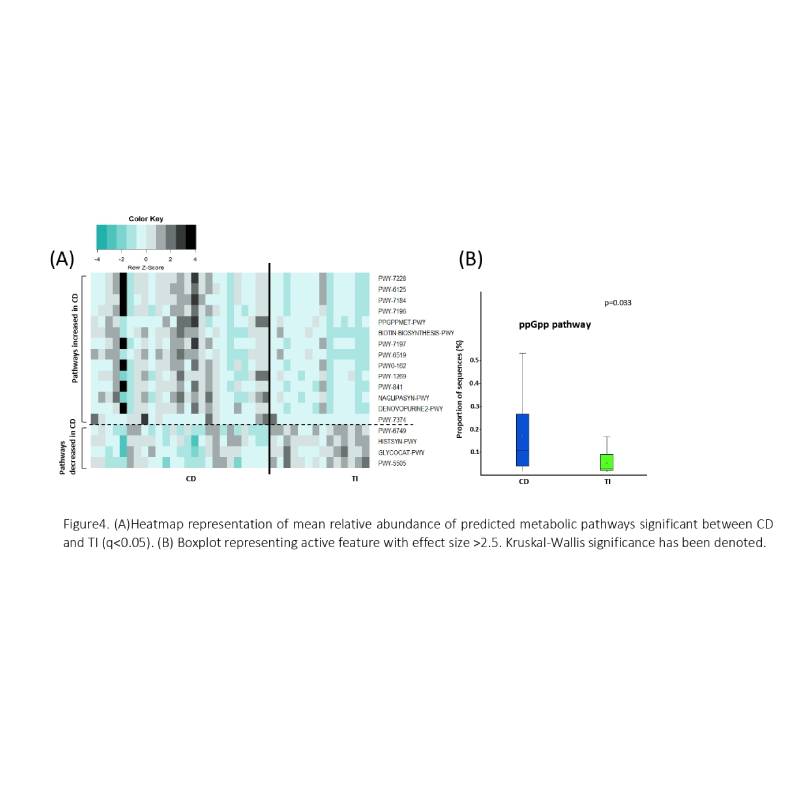
Microbiome data analysis revealed a progressive decline in α-diversity(p=0.002) in CD and different β-diversity (Bray-Curtis dissimilarity p=0.001) compared to TI and healthy(Fig1). Overall Firmicutes were lower(p=9.81e-4) while Desulfobacterota were higher(p=8.65e-3) in CD than TI(Fig2A). Faecalibacterium prausnitzii (p=0.04) and Roseburia inulinivorans (p=0.03) were markedly reduced while Parabacteroides distasonis (p=9.91e-3) and Bilophila wadsworthia (p=7.36e-3) were elevated in CD (Fig3). A microbial signature covering 8 non-overlapping taxa was deduced to distinguish CD and TI (3 taxa reduced, 5 elevated in CD compared to TI) (Fig2B). PICRUSt analysis indicated 18 significant metabolic pathways differentiating CD and TI. Bacterial stress alarmone PPGPP-MET was significantly higher in CD (p=2.04e-3) (Fig4). FCal was significantly higher in CD with mean value (±SD) as 549.12±390.4 and 167.15±191.8µg/g in TI (p=0.002).


Conclusion
A distinct gut microbiome profile was observed in TI compared to CD. Exploitation of these differences can potentiate differential diagnosis of CD and TI. Further research on the spectrum of dysbiosis is warranted. This may help in differentiating patients with early-CD who require active therapy from those with non-specific TI ulcers that resolve over time.


